Unlocking mRNA Therapeutics: Cell-Free DNA Synthesis Is Powering the AI Era of Genetic Medicine
Cell-free gene synthesis, AI-powered design and advanced mRNA delivery are ushering in a new era of genetic medicine.
The COVID-19 pandemic thrust messenger RNA (mRNA) into the spotlight, underscoring its promise as a nimble, programmable platform for delivering genetic instructions directly to cells. The success of mRNA vaccines – rooted in years of scientific groundwork – has expanded mRNA’s role into broader therapeutic applications, including protein replacement, genome editing and cell and gene therapies (Figure 1). These modalities are being investigated across diverse indications, from cancer to autoimmune and rare genetic diseases.¹ This breakthrough is creating opportunities to replace conventional treatment approaches that are slower, more expensive and more complex to manufacture.
Yet realizing the full potential of mRNA therapies requires more than a compelling concept. It demands unprecedented speed, accuracy and adaptability in the way researchers design and manufacture genetic payloads. Accelerating iterative design cycles required for optimal performance, as well as streamlining the process to scale production, is imperative.
mRNA: A programmable therapeutic platform
At its core, mRNA’s power comes from two defining traits: its programmability and its role as a versatile platform technology. Proteins can be changed simply by altering the nucleotide sequence, while the backbone, delivery vehicle and manufacturing process remain largely constant. Once validated, the platform allows new therapies to advance without starting from scratch, dramatically shortening development timelines and simplifying the regulatory process.
mRNA-based vaccines exemplify platform agility – they can be rapidly designed, tested and refined without the labor-intensive protein engineering required for traditional approaches. During the COVID-19 pandemic, this flexibility was critical to developing vaccines in under a year and enabling swift updates as new variants emerged. The same speed and programmability underpins personalized cancer vaccines, where each formulation is tailored to a patient’s unique neoantigen sequence. To stay ahead of tumor evolution, these therapies must be produced within weeks – marking a shift from one-size-fits-all treatments to on-demand personalized medicine.
Beyond vaccines, mRNA unlocks entirely new therapeutic strategies. Conventional cell and gene therapies, such as CAR T or genome-edited treatments, depend on ex vivo methods that are costly and time-intensive. In contrast, mRNA can be delivered in vivo via nanoparticles, enabling cells to be engineered or diseases treated directly within the body – bypassing many of the manufacturing and quality control bottlenecks of traditional approaches.
With its programmability and platform approach, mRNA stands as one of the most versatile therapeutic technologies to date.¹˒² Clinical trials are underway for mRNA therapies targeting melanoma, cystic fibrosis and autoimmune diseases such as multiple sclerosis. As the global biopharmaceutical market is projected to exceed $1 trillion by 2032,3 mRNA is poised to become a foundational pillar of next-generation medicine.
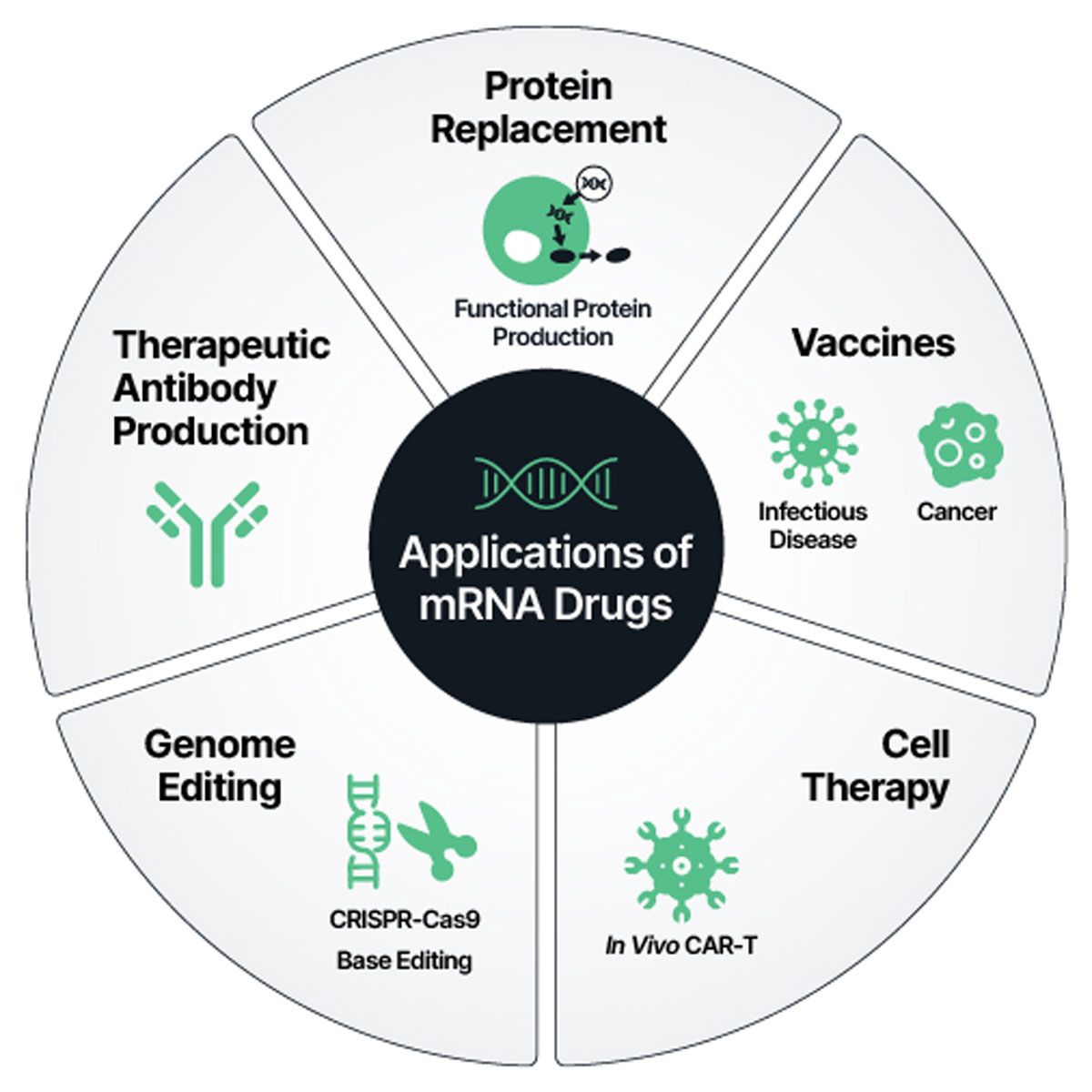
Figure 1: mRNA is a versatile therapeutic that can be used for diverse applications like replacing proteins, constructing cell therapies and engineering the genome. Credit: Elegen.
Challenges with traditional DNA templates for IVT
Despite its promise, mRNA’s emergence as a versatile platform for advanced therapeutics is limited by several bottlenecks. Chief among them is the in vitro transcription (IVT) process, where synthetic DNA templates serve as blueprints for mRNA synthesis. During IVT, RNA polymerase transcribes a linearized DNA template into mRNA, with a 5′ cap and poly(A) tail – critical for stability and translation – added either during or after transcription.
Traditionally, IVT templates are produced through bacterial cloning, a foundational yet inefficient approach. Working with cells introduces the risk of contamination from host-derived components such as endotoxins, genomic DNA and enzymes, which must be removed to avoid immune activation in downstream applications.
Another major challenge is maintaining consistent poly(A) tails, a key determinant of mRNA stability and translational efficiency. The tail can be added post-transcriptionally to the mRNA, incorporated via PCR into the DNA template or encoded directly in the DNA. Each method has trade-offs: post-transcriptional tailing and PCR-based approaches can introduce polymerase-driven variability, while encoding the tail directly in DNA avoids this, but long homopolymeric tracts are prone to bacterial recombination and instability. Such disruptions can compromise DNA integrity and produce heterogeneous mRNA.
Finally, cell-based production of DNA templates constrains development timelines. As demand for faster and more affordable DNA manufacturing grows, bacterial cloning workflows remain slow and labor-intensive.4 Bacterial transformation, colony screening, amplification and endotoxin removal extend turnaround times and complicate scale-up, significantly slowing mRNA discovery and development.
Cell-free DNA synthesis: Speed and precision without the baggage
To overcome these challenges, contract development and manufacturing organizations and mRNA manufacturers are increasingly adopting cell-free gene synthesis. Streamlined, automation-friendly cell-free workflows enable rapid generation of DNA templates. By eliminating the use of cells, these processes bypass risks of contamination and bioburden, reducing the need for extensive QC testing and dramatically accelerating production.
Cell-free synthesis also minimizes variability in the resulting mRNA. For instance, encoding a poly(A) tail directly into plasmid templates can lead to recombination-driven differences in tail length. In contrast, encoding the tail directly into cell-free templates improves stability and removes the need for downstream tailing, yielding more consistent and reliable mRNA products – a critical need in therapeutic applications.5
The role of AI in accelerating mRNA development
AI is ushering in a new era of rational, data-driven design for mRNA therapeutics, creating a continuous feedback loop between design, synthesis and testing.6 Its most immediate impact is in the discovery phase, where AI accelerates the design and optimization of candidate sequences. After a target is selected, designs are generated in silico, synthesized as DNA templates and transcribed in vitro for evaluation. The resulting mRNA is functionally tested in cell culture for expression, stability and toxicity, with the data feeding back into AI models that propose optimized sequence variants. Machine learning and neural network–based tools are already proving highly effective at predicting and improving sequence stability, translational efficiency and immunogenicity.7,8,9 The new variants often require rapid, de novo synthesis to keep projects on track. Here, cell-free DNA synthesis offers a critical advantage: long, complex sequences can be produced quickly and with a stable, encoded poly(A) tail (Figure 2).
The ability to rapidly produce diverse templates on demand is a game-changer for mRNA design and production. Researchers can initiate transcription with minimal delay, optimize candidates quickly and advance therapies at unprecedented speed. By combining AI-driven design with cell-free DNA synthesis, developers can test more variants in parallel, identify top performers faster and refine them iteratively – cutting the time and cost of traditional trial-and-error workflows.
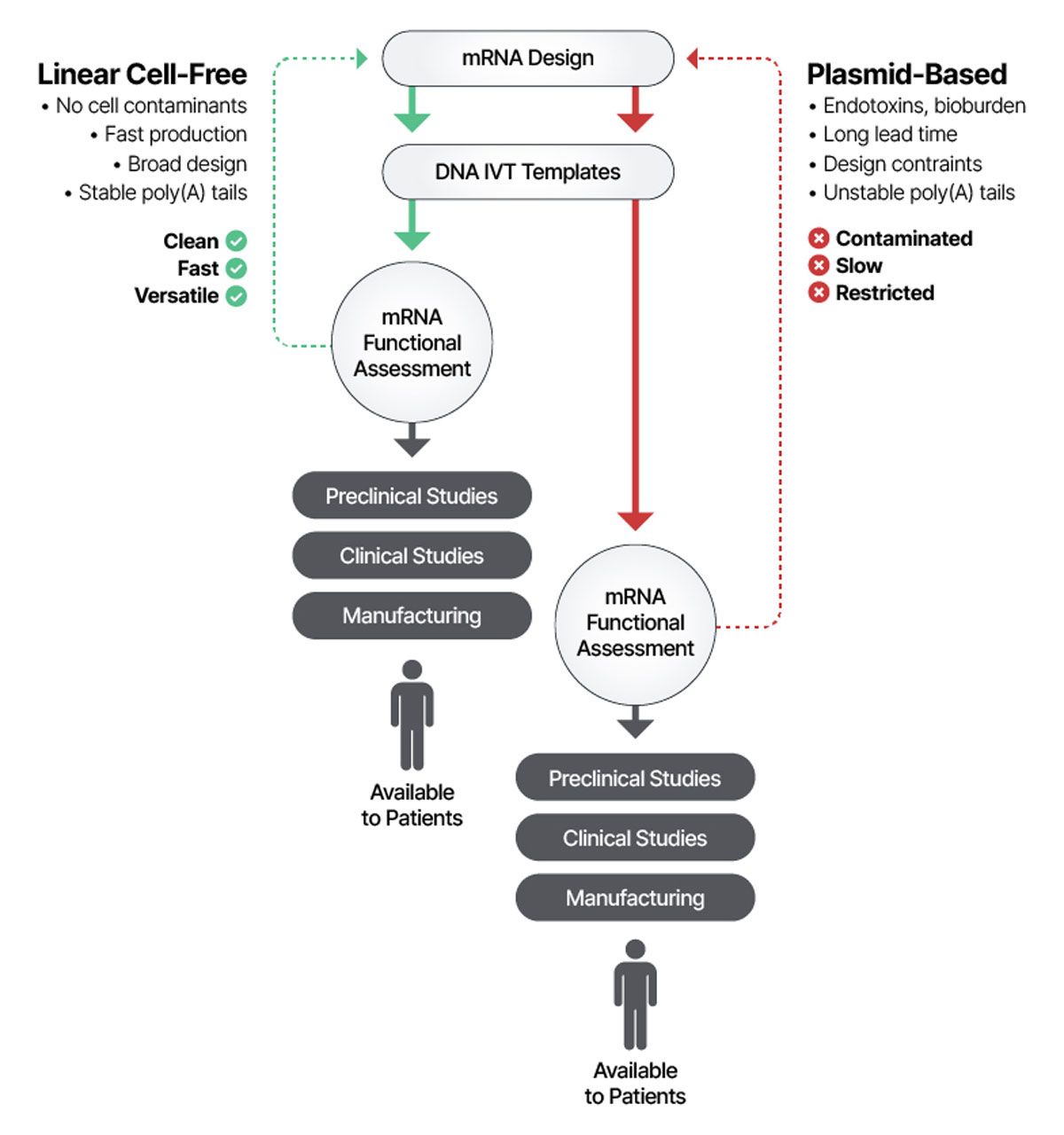
Figure 2: Cell-free linear IVT templates enable rapid testing, optimization and advancement of mRNA candidates, accelerating new therapies to patients while avoiding the delays, contamination risks and design limitations of plasmid-based templates. Credit: Elegen.
Optimizing IVT workflows and scaling up
As mRNA candidates advance, scale-up and Good Manufacturing Practice compliance become critical. By eliminating the need for cell culture, cell-free synthesis offers a more reliable, cleaner and automation-friendly process. It also allows a smaller manufacturing footprint and greater capital efficiency. Together, these advantages create a simpler, more robust workflow that aligns more easily with Chemistry, Manufacturing, and Controls requirements for therapeutic development.
The road ahead: Programmable, personalized and scalable therapeutics
The convergence of cell-free gene synthesis, AI-powered design and advanced mRNA delivery is ushering in a new era of genetic medicine. Together, these technologies are reshaping what’s possible in the design and deployment of programmable therapeutics.
As advanced computational tools uncover increasingly complex and diverse sequence landscapes, the challenge is shifting – from determining what mRNA to test, to determining how quickly the corresponding DNA templates can be produced. Today, developers often rely on multiple suppliers to obtain the sequence variants proposed by their AI models. What’s needed instead are one-stop shops capable of rapidly designing and producing these templates on demand.
As manufacturers work toward resolving the DNA synthesis bottleneck, mRNA therapeutics will expand into new frontiers – from personalized cancer vaccines and rapid-response platforms for emerging pathogens to treatments for rare genetic disorders. The ability to design, synthesize and validate therapies in a fraction of the time once required will be essential to keeping pace with evolving clinical needs.
Equally important, the scalability of cell-free workflows ensures that these innovations can reach the market faster, more cost-effectively and with greater flexibility. As regulatory frameworks for mRNA therapeutics continue to mature, such capabilities will be central to streamlining approval processes and ensuring consistent product quality across research and clinical applications.
Conclusion
mRNA has proven itself to be more than a temporary solution to a global crisis. It is a foundational platform for the future of medicine. But to fully unlock its potential, we must overcome the production and design constraints of legacy technologies.
Cell-free DNA synthesis and AI-driven sequence engineering are enabling precisely that. By eliminating bottlenecks, increasing design freedom and enabling scalable production, these innovations are accelerating the pace of therapeutic discovery and delivery.
The future of mRNA therapeutics is fast, programmable and highly scalable – and it’s being built today.



