Proteomics is transforming the way researchers approach biomarker discovery, drug development and precision medicine, providing insights into protein dynamics at scale.
Yet, conventional methods for analyzing protein differences across populations are often labor-intensive and technically demanding, limiting their application in large studies. There is a clear need for approaches that make proteomic analysis more scalable, accessible and clinically relevant.
This article explores how NGS-based proteomics addresses these challenges, highlights recent advancements in the field and examines its impact on multiomics research and precision medicine.
Download this article to discover:
- How NGS-based proteomics is advancing the field of precision medicine
- The role of proteomics in multiomics research through real-world case studies
- NGS-based proteomics solutions that make multiomics research more accessible, scalable and insightful
Article
How NGS-Based Proteomics Is
Advancing Disease Insights and
Precision Medicine
Introduction
For decades, scientists have studied genetic predispositions to
better understand why diseases develop, when they emerge and
how they progress. Precision medicine recognizes people’s
genetic differences and creates prevention and treatment plans
tailored to their unique biology. However, DNA is only part of the
story, as our genetic code provides instructions for making
proteins. These proteins are responsible for carrying out nearly
every biological process in the body, from metabolizing sugars to
regulating immune responses.
Genetic differences can lead to variations in the quantity, structure
and function of proteins. For example, a genetic variant might
affect glucose metabolism, increasing the risk of developing
type 2 diabetes. However, knowing that a person carries a
particular variant doesn’t always reveal the full picture, as it
doesn’t indicate how much of the protein is being produced or
how well it is functioning.
Proteomics addresses this gap by directly quantifying the proteins
and revealing how genetic variation translates into biological
activity. Unlike genomic data, which reflects potential risk,
proteomic analysis captures dynamic, real-time physiological
processes occurring in the body.
In this article, Dr. Cindy Lawley, Senior Director of Global
Population Health at Olink Proteomics, discusses the application
of next-generation sequencing (NGS)-based proteomics to
advance disease insights, personalize therapeutic journeys and
improve disease risk prediction.
Watch the complete Teach Me in 10 episode with Dr. Cindy
Lawley here.
Learn more about Olink’s NGS-based proteomics here .
How NGS-based proteomics works
NGS-based proteomics enables large-scale protein analysis and
has only recently become advanced enough to perform
proteomics profiling across large populations.
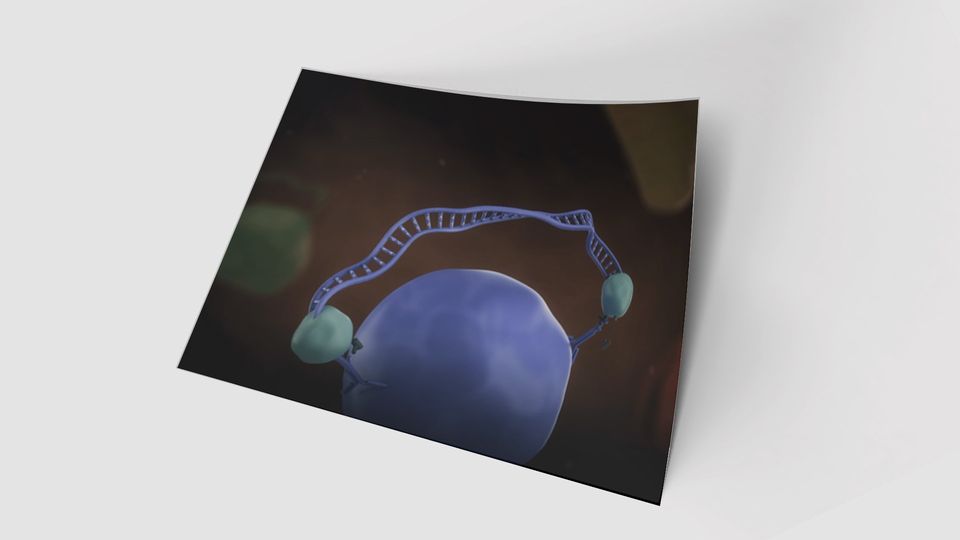
One approach driving these advances is the Proximity Extension
Assay (PEA). The PEA technology involves two antibodies, each
tagged with a single DNA strand, that bind specifically to their
target protein. When both antibodies attach to the same protein,
their DNA tags hybridize and are extended to form a complete
sequence. This DNA ‘barcode’ is an identifier for the protein in
question, which can be detected and quantified using NGS
(Figure 1). This process effectively converts a protein signal into
a measurable DNA signal, with the number of DNA tags
accurately reflecting the protein concentration in the sample.
This high-throughput method enables the rapid, precise and
large-scale measurement of thousands of proteins in as many
samples. This data helps researchers identify protein biomarker
patterns that may support more accurate disease prediction and
therapeutic development.2
The UK Biobank: A proteomics
treasure trove
To uncover population-wide protein biomarker patterns and gain
deeper insights into disease development and progression,
high-throughput NGS-based proteomics is used to generate
proteomics data from large-scale biobank samples. The UK
Biobank is one of the world’s most valuable research resources,
comprising biological samples and health data from around
600,000 participants who enrolled in the early 2000s. Initially, the
UK Biobank collected genetic data on all participants, making it
possible to study inherited disease risk. Later, the UK Biobank
Pharma Proteomics Project (UKB-PPP) expanded on this by
including protein biomarker profiling in the database, effectively
combining genetic, proteomic and health data in a single resource.
The pilot phase of the UKB-PPP was a collaboration between 14
pharmaceutical and biotech companies that characterized the
plasma proteomic profiles of over 54,000 UK Biobank
participants.1 The study demonstrated how protein-based risk
scores – a likelihood prediction of developing a certain disease
– could significantly outperform genetic risk scores alone for
predicting many diseases. The study has been accessed over
168,000 times and cited around 700 times, reflecting its global
impact. This flagship paper has laid the groundwork for numerous
studies worldwide, establishing the UK Biobank as a vital
international resource for advancing health research and precision
medicine. Building on the success of the pilot phase, it was
announced in January 2025 that Olink® Explore HT (covering over
5,400 proteins) would be used to profile all 500,000 UK Biobank
participants. This expanded dataset promises to reveal even more
about the intricate connections between proteins and disease,
laying the groundwork for targeted therapies and enhanced
diagnostic tools.
How research takes a concept
to the clinic
Several research groups have conducted studies to explore the
utility and validity of a proteomics approach for disease
prediction. Many studies involve modeling how protein signals
perform compared to genetic signals. For example, a research
team built predictive models for 23 different diseases by
combining multiple protein measurements into a single score
(ProteinScores), estimating a person’s risk of developing a disease
within 10 years. The team found that, for conditions such as type 2
diabetes, this score significantly improved prediction accuracy
beyond traditional risk factors, lifestyle variables and even clinical
and genetic biomarkers.2
Figure 1. Proximity extension assay (PEA) technology, whereby: 1) antibodies with DNA tags bind to the target
protein, 2 and 3) the DNA tags hybridize when pairs are correctly matched and 4) a unique DNA barcode is
created for each protein. This DNA tag is then quantified using NGS.3
NGS-based proteomics can also be used to predict cancer
development. In another study, UK Biobank data was used to
develop protein signatures that could predict 19 blood and solid
tumor cancers as early as 12 years before clinical diagnosis.3
Beyond cancer, a similar approach was used for 67 common and
rare diseases, where protein scores built from as few as 5 to 20
proteins outperformed current clinical tools for predicting who
will develop these conditions.4
Together, these findings highlight the potential of proteomic
profiling to enable earlier and more accurate disease risk
prediction across a wide range of conditions.
Yet, further clinical validation is required to be able to apply this
approach in routine clinical diagnostics and patient care. One such
example is the development of a multiple sclerosis test based on
protein signatures. Beginning with the identification of 1,416
related proteins in a 2017 discovery study, several research teams
were able to validate 18 proteins as part of a dashboard for
monitoring the condition (Figure 2).5,6,7,8 By monitoring these
indicative proteins, a personalized medication program can be
made for each patient, supporting their therapeutic journey as
they receive various medications throughout their life.
NGS-based proteomics: A clinical
crystal ball?
NGS-based proteomics offers a powerful new tool for predicting
disease risk and guiding personalized medicine. By capturing
thousands of proteins at scale, researchers can see beyond
genetic predisposition to the intertwining biological processes
driving disease. This approach enables researchers to predict
disease development several years before symptoms arise, as
shown in studies predicting diabetes, cancer and
neurodegenerative conditions.2,3,4,5,6,7,8
Combined with genetic approaches, proteomics allows
researchers to understand not only inherited risk but also lifestyle
and environmental factors, making it an exceptionally rich source
of insight for precision medicine. Currently, validated clinical tools
are translating these discoveries into real-world care, supporting
treatment decisions tailored to individual patients.
As large-scale resources like the UK Biobank continue to integrate
genetic, proteomic and health data, the potential for discovering
new biomarkers and refining risk scores will only grow. While
challenges remain in clinical validation and implementation,
NGS-based proteomics is poised to become a cornerstone of early
detection, risk stratification and personalized treatment planning.
By promoting this technology, the healthcare sector can move
toward a future that is truly predictive, precise and personalized,
giving clinicians a new tool that may amount to a molecular-level
crystal ball for patient care.
Learn more about Olink’s NGS-based proteomics here .
Dr. Cindy Lawley, Senior Director of
Global Population Health at Olink
Proteomics, part of Thermo Fisher
Scientific, has helped develop
solutions to better understand
genetic risk in diverse populations
and holds several Excellence in
Technology Transfer awards for her work. She currently
drives population health initiatives, working closely with
large cohort studies.
Figure 2. The development pathway of the multiple sclerosis disease activity (MSDA) test.
Discovery Study
2017
Panel Development
2018
Analytical Validation
2020
Clinical Validation
2023
300 Samples 650 Samples >3,000 Samples 600 Samples
1,416
Proteins
21
Proteins
18-plex
MSDA Test4
References
1. Sun BB, Chiou J, Traylor M, et al. Plasma proteomic associations with genetics and health in the UK Biobank.
Nature. 2023;622(7982):329–338. doi:10.1038/s41586-023-06592-6
2. Gadd DA, Hillary RF, Kuncheva Z, et al. Blood protein assessment of leading incident diseases and mortality in
the UK Biobank. Nat Aging. 2024;4(7):939–948. doi:10.1038/s43587-024-00655-7
3. Papier K, Atkins JR, Tong TYN, et al. Identifying proteomic risk factors for cancer using prospective and exome
analyses of 1463 circulating proteins and risk of 19 cancers in the UK Biobank. Nat Commun. 2024;15(1):4010.
doi:10.1038/s41467-024-48017-6
4. Carrasco-Zanini J, Pietzner M, Davitte J, et al. Proteomic signatures improve risk prediction for common and
rare diseases. Nat Med. 2024;30(9):2489–2498. doi:10.1038/s41591-024-03142-z
5. Qureshi F, Hu W, Loh L, et al. Analytical validation of a multi‐protein, serum‐based assay for disease activity
assessments in multiple sclerosis. Proteomics Clin Appl. 2023;17(3):2200018. doi:10.1002/prca.202200018
6. Chitnis T, Foley J, Ionete C, et al. Clinical validation of a multi-protein, serum-based assay for disease activity
assessments in multiple sclerosis. Clin Immunol. 2023;253:109688. doi:10.1016/j.clim.2023.109688
7. Sanchez A, Sheng E, Eagleman S, et al. Real-world clinical utility of a multi-protein, blood-based biomarker
assay for disease activity assessments in multiple sclerosis. Mult Scler J Exp Transl Clin. 2025;11(2).
doi:10.1177/20552173251331030
8. Chitnis T, Qureshi F, Gehman VM, et al. Inflammatory and neurodegenerative serum protein biomarkers
increase sensitivity to detect clinical and radiographic disease activity in multiple sclerosis. Nat Commun.
2024;15:4297. doi:10.1038/s41467-024-48602-9
www.olink.com
Olink products and services are For Research Use Only and not for Use in Diagnostic Procedures.
This document is not intended to convey any warranties, representations and/or recommendations of any kind,
unless such warranties, representations and/or recommendations are explicitly stated.
Olink assumes no liability arising from a prospective reader’s actions based on this document.
OLINK and the Olink logotype are trademarks registered, or pending registration, by Olink Proteomics AB.
© 2025 Olink Proteomics AB. All third-party trademarks are the property of their respective owners.
Olink products and assay methods are covered by several patents and patent applications https://www.olink.com/patents/.
Olink Proteomics, Dag Hammarskjölds väg 52B , SE-752 37 Uppsala, Sweden
1146, v2.0, 2022-10-1